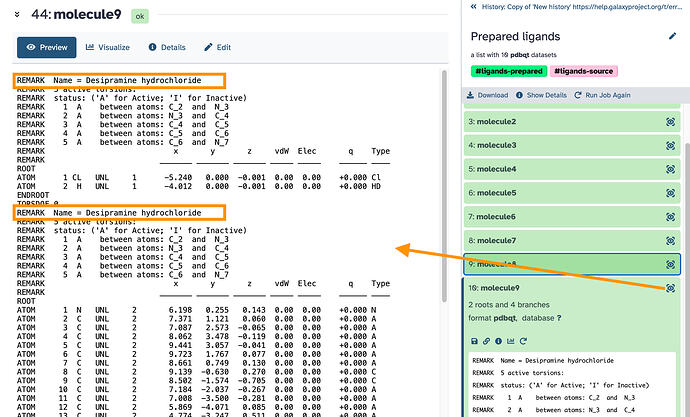
I’m encountering an error while running a molecular docking simulation. I’ve attached the full error message below. Could someone please help me understand what’s causing the issue and how to resolve it?
Thank you for your time and support
Tool ID -toolshed.g2.bx.psu.edu/repos/bgruening/autodock_vina/docking/1.2.3+galaxy0
*** Open Babel Warning in ReadUnimplementedBlock
COLLECTION blocks are not currently implemented and their contents are ignored.
1123 molecules converted
1123 files output. The first is ligand1.pdbqt
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Atom type B is not a valid AutoDock type (atom types are case-sensitive).
> ATOM 1 B UNL 1 -1.214 -2.726 0.000 0.00 0.00 +0.000 B
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Atom type B is not a valid AutoDock type (atom types are case-sensitive).
> ATOM 11 B UNL 1 0.750 -9.093 0.000 0.00 0.00 +0.000 B
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
=================
..
s in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Atom type Au is not a valid AutoDock type (atom types are case-sensitive).
> ATOM 9 AU UNL 1 3.750 -1.299 0.000 0.00 0.00 +0.000 Au
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
==============================
*** Open Babel Warning in PerceiveBondOrders
Failed to kekulize aromatic bonds in OBMol::PerceiveBondOrders (title is =)
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
==============================
*** Open Babel Error in TetStereoToWedgeHash
Failed to set stereochemistry as unable to find an available bond
PDBQT parsing error: Unknown or inappropriate tag found in flex residue or ligand.
> ROOT