hi,
im trying to run DEseq2 on 2 file. one for control and one for treated. but it showing an error that input file has repeated input file.
can anyone help me?
Replicates are required with this tool plus most DE tools.
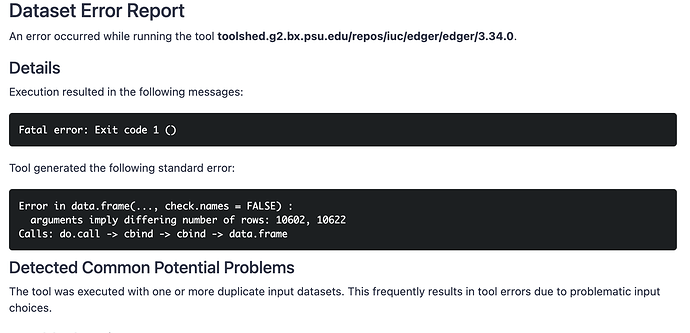
The “different number of rows” report indicates that the same reference annotation was probably not used for each counting step.
FAQ: Help for Differential Expression Analysis
Tutorials were already referenced above. Plus I added a few tags to your post that will likely help.
Best
I did tried with replicates and still having the same issues.
If you have replicates now, check that the number of lines for each count dataset are the same. There should be at least four unique count datasets input to the tool.
Each line represents a gene (plus optionally, one header line). Should the number of lines be different between count files, then that indicates a problem with the reference annotation or perhaps how you are counting (this tool expects counts by “gene”).
The FAQ above covers nearly all the issues that can come up with this tool and other similar tools. Your original posted error had two problems: 1) no replicates and 2) a different number of lines (genes) for each of the two inputs.
Check the new error message: If the line counts are different, you’ll need to review the upstream analysis to fix the annotation/counting problems. The “End to End” tutorials have an example. Galaxy Training!
Ps: Please help to keep the conversation on a single topic by avoiding cross-posting to other topics. I saw that you posted on the topic below about this same issue. That post is a good reference, specifically the sentence below, but let’s keep troubleshooting your issue in this topic. If you need or want to reference other topics, capture the link and post it back here.
Hi,
I used the same annotation for all of them but i have different number of rows for replicates. How can i solve it? I followed the steps from above steps you mentioned but still having the same issues.
This shouldn’t be possible unless some kind of manipulation was done after counting.
Are you working at a public Galaxy server? Which one (URL)?
Yes im working on public server usegalaxy.org. i used dm6 reference annotated file in gtf format for all of my analysis. I didn’t do any manipulation after stringtie and did feature count also and then did deseq2.
hi,
anyone can help on that?
Please submit the problem from the error dataset and we can help with a closer review. How to: click on the “bug” icon from the red error dataset, fill out the form adding comments, and submit.
Please be sure to:
- Leave all upstream steps/datasets in the history as undeleted.
- Capture the URL from this topic/conversation and paste that into the comments, so we can link the two together.
Thanks!