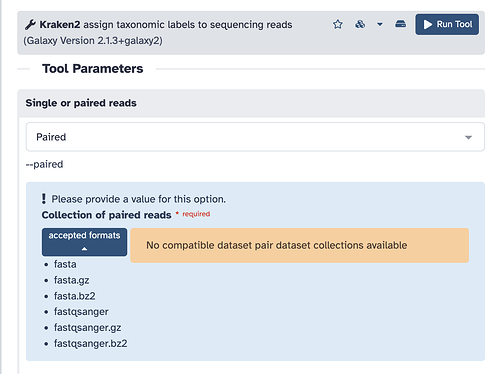
Hi, I have a set of paired-end samples generated from the Illumina platform. I’ve been trying to run Kraken2 on them using the Galaxy Europe server with the suggested formats. However, I noticed that paired-end input doesn’t seem to be supported when I try to select the samples.
Welcome @raptor_007
This is happening with the input setting on the Kraken2 tool form? Like this?
If so, one of these is likely the problem:
-
The input reads are not in the active history. You can try refreshing your browser window, or clicking on the Home Galaxy icon in the top left corner, then switching to the history, before loading the tool form up again. Hands-on: Understanding Galaxy history system / Understanding Galaxy history system / Using Galaxy and Managing your Data
-
The reads are not assigned one of the supported datatype “accepted formats”. Illumina reads would be fastqsanger or fastqsangergz. If the reads do not have that datatype from the start, then you’ll need to correct it (maybe by reloading the data) then redoing the upstream QA steps (since these also also rely on the correct quality score scaling definition in the datatype). Getting Data into Galaxy
-
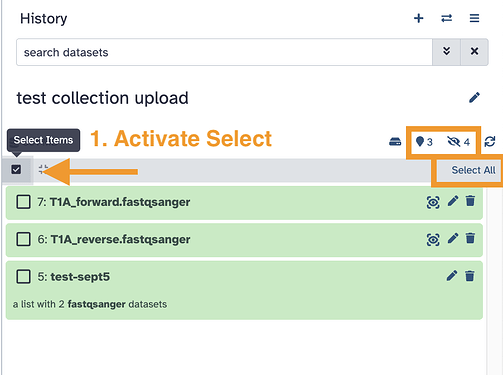
Your data is not in a collection yet. This tool expects a Collection of paired reads for the shape of the data, not individual files. FAQ: Datasets versus collections. Load your data up using the Collection tab in the Upload tool or create it after from data already in your history.
Screenshots for the Collection folder building functions
Resources
We have several tutorials for Kraken2 if you would like to work with examples, or maybe find a workflow to get you started!
 GTN tutorials for Kraken2: assign taxonomic labels to sequencing reads
GTN tutorials for Kraken2: assign taxonomic labels to sequencing reads- Search the Public Workflows tab at any public Galaxy server for more workflows!
- And, production workflows from the IWC can be found at → https://iwc.galaxyproject.org/
Hope this helps but please let us know if it actually does or if you need more help. You can share screenshots or your history for more context! ![]()