Dear support team at GALAXY,
I am trying to perform a RNA-seq analysis for D. melanogaster. I did the mapping with Bowtie2, using the genome assembly: dmelo_r6.32 and all were fine, nice mapping stats etc. Then, I tried to generate the necessary matrices for the DEseq tool via using the tool HTseq:
When I used the corresponding .GFF ( dmel-all-r6.32.gff.gz ) for this particular assembly, I got the error below:
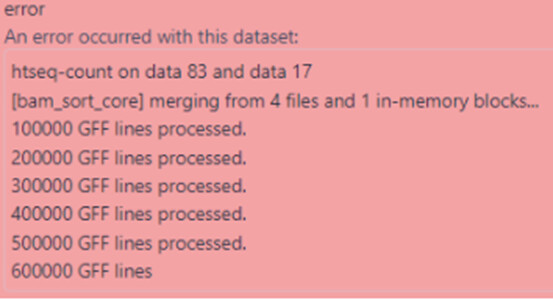
When I used the .GFF for the assembly 6 from the NCBI, then I got the error below:
Any input on how to overcome this obstacle in my analysis will be highly appreciated.
Manolis
