I want to summarize all fastqc results from 2 pair-end samples (so in general 4 collections) in one MultiQC file. I receive the output file however it returns only one collection summary instead of 4. I tried flattening collection before puting them into MultiQC, sadly it didn’t work.
Hello @Orange_Pomeranian
Yes, MultiQC is very picky and does some automatic interpretations of the sample file names that isn’t super obvious. This happens everywhere, not just Galaxy, so you’ll find lots of discussion online with a search like “multiqc missing samples”.
I have a demonstration workflow that performs this correctly. Would you like to compare to see what I did?
- Flattening before FastQC is the import part, since that avoid the “duplicated” sample names issues.
- You’ll also want to run through a step to rename the element identifiers in either your “original” OR “trimmed” read collections to avoid a conflict at that level. I did the latter.
- A bit complicated! But you’ll see what I did and understand better with the example.
You can also search at the UseGalaxy servers with that same term under the Activity bar → Workflows → Public workflows tab to find it. If you are working someone else, you can still review it in context. Download a copy, then import at your other server.
Hope this helps! ![]()
Try replying again now.
And a note: it is hard to tell what is going on in a workflow without seeing the actual shared workflow. The graph is just one part of it. See my example for the renaming portions. The graphic in the direct message was missing some steps compared to the one I shared – in particular, the element renaming steps. Be sure to notice how I applied those names – the common ways people may rename will be part of what I meant here:
does some automatic interpretations of the sample file names that isn’t super obvious
Example: “trim” added on will get removed, but something like “trimx” will pass through.
How this works is how it would work if these files were all in folders on your computer that are run through the tool, so this is not Galaxy specific, it is part of how the underlying tools parse data to be “helpful”.
Also, be sure to run my workflow on at least two samples so you can see how the results come out. The hidden files will contain all of the intermediate steps in action.
From here, please try to reply again and to include shared data for review. The input + output data along with the workflow runs is a good idea. So, that could be two shared workflow invocations: one for my workflow, and one for yours.
Thanks!
Hi below I am uploading the main workflow to which i want to add all 4 outputs from fastqc.
heres the modified version that sadly didn’t work as i expected
Thanks for sharing these! I see the problem.
Notice that you have used
- Upzip collection - to get the forward and reverse reads into different list collections.
- Flatten collection - to transform a paired end collection organized by sample into a flat list with both forward and reverse reads together, but with unique full sample names: base sample name plus forward/reverse designation combined into the element identifier.
It is fine to keep the forward and reverse reads together. This means you can run just 2 above and skip using 1.
Then, to avoid conflicts between the trimmed and untrimmed, you can modify the element identifiers for one of the versions of the collections, or you can modify both and let the tools sort all of the the data by an “inferred sample” name.
How this works: the base name in all of the different collection identifiers is the “sample name” – specifically, the “sample” is the part of the element identifier before the first underscore _ character. Content after the first _ is also interpreted but you can control how this is handled.
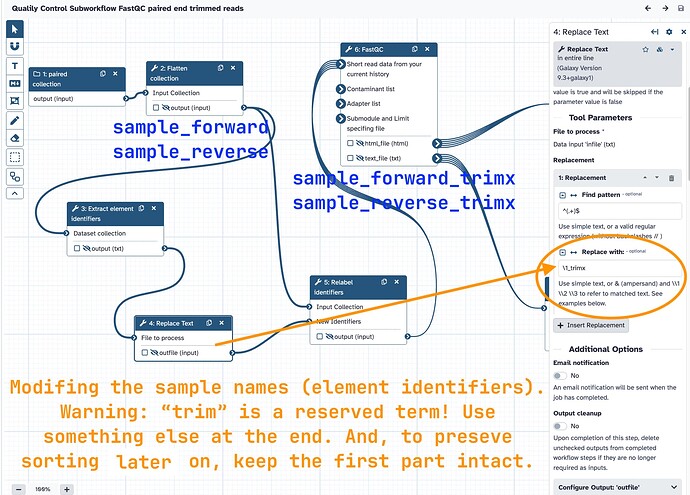
Warning! These tools have some reserved terms!! If you only add on _trim or _trimmed, it will be parsed out by FastQC and you’ll lose samples in the report! So add in something else. I used _raw and _trimx. Later on, MultiQC still parsed out _raw in the final report, but I was Ok with that. You could experiment with different terms to see what passes through or not.
_rawwas added for the raw reads sample names_trimxwas added for the trimmed reads sample names
You could use something like _banana and _apple and that would probably work too! ![]()
Screenshots
Your workflow with the conflict annotated.
One of my workflow’s subworkflows annotated. I did almost the same thing for the raw reads in the other subworkflow (used a different term at the end _raw).
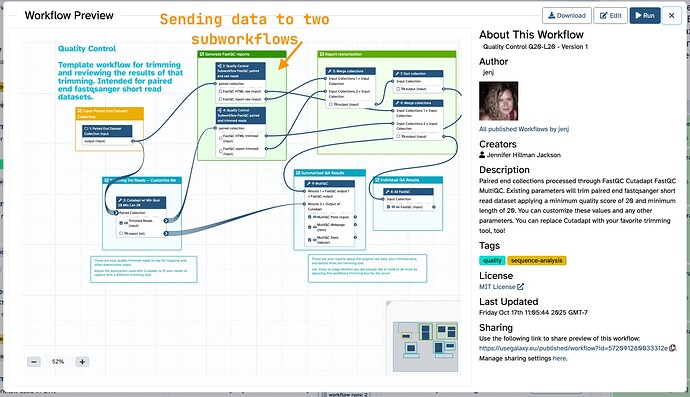
My main workflow. The subworkflows consume a paired collection then output the FastQC reports. All four versions of the reports have a unique full sample name, but have the base name the same! The results were returned, then sorted, then summarized by MultiQC.
I moved these over to the EU server (from the ORG server) to make it easier to find and examine in more places. Search the Activity bar → Workflows → Public Workflows tab with the keyword “quality”. Or, these links will take you to each directly.
- Main workflow → https://usegalaxy.eu/u/jenj/w/quality-control-q20-l20
- Subworkflow for raw reads passing through FastQC → https://usegalaxy.eu/u/jenj/w/qualily-control-subworkflow-fastqc-paired-end-raw-reads
- Subworkflow for trimmed reads passing through FastQC → https://usegalaxy.eu/u/jenj/w/qualily-control-subworkflow-fastqc-paired-end-trimmed-reads
The subworkflows can be used by anyone in other workflows. They consume a paired end collection of Illumina reads and output a collection of FastQC reports. Which trimming tool you used won’t matter. You can also import and edit them any way you want! Just give them a unique name to better help to keep track of what is doing what, and add yourself as an editor and a short description of what you changed if you decide to publish on the server.
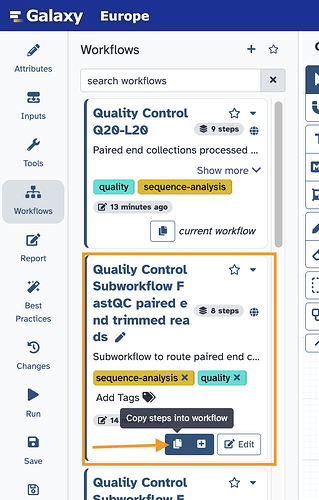
How to use a subworkflow? Import it first, make changes if you want to (change the _trimx to be something else if you want), save, then go in to edit your primary workflow.
Once in your primary workflow, you can add in a subworkflow’s content to your workflow. You can just copy the steps, or you can keep the steps broken out into a subworkflow.
Unique sample names are required when using the FastQC/MultiQC tools directly on the command line, too. People will go in an rename files in their sequence directories to avoid the conflicts. You will be doing the same thing in Galaxy – the element identifiers of collections are analogous to the “file names” that tool’s interpret in this context.
So, you have some choices! Hope this helps but let’s know if it actually does and if you are able to get this to work! You don’t have to use my workflows – but examining what I did will probably help. ![]()
Great thanks, it finally worked ![]()