I’ve been having errors using stringtie with Deseq2. After I have my HISAT BAM files I put them into Stringtie with genecodevM31>stringtie merge> stringtie again using the merge file as the reference file then I obtain gene count files. However, my gene count files have different number of rows for my different samples and not the same. In my previous files I had no trouble but this round all my row values are different.
Welcome, @rjsoh
The Stringtie form has a default option to predict new gene models. You’ve already done that with the first round, and merged those together to create a new “reference”. Now you need to set the option to only use the existing reference data for the final gene counting round. This generates count that are all based on the same annotation, without more predictions, and all count files will then have the same number of rows (same set of genes).
Choose your merged GTF file, then find the option here on the form, and toggle it to Yes.
Xref StringTie error generating gene and transcripts counts files
Hope this helps! ![]()
Hello,
Thanks so much for this info. I have been using the stringtie merged as the reference but the resulting gene counts is still different between my replicates and my samples…
Hi @rjsoh Odd … do you want to share your history? It is hard to guess more about what else might be going wrong.
Tutorial example of how to create a master reference annotation – this specific step → https://training.galaxyproject.org/topics/transcriptomics/tutorials/de-novo/tutorial.html#transcriptome-assembly
And, an alternative way to do the counting – at this step → https://training.galaxyproject.org/topics/transcriptomics/tutorials/de-novo/tutorial.html#analysis-of-the-differential-gene-expression
Thanks so much for your help,
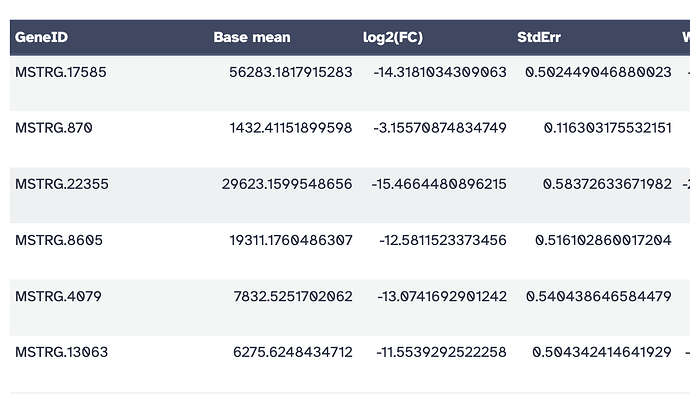
I tried doing the counting through featurecounts and GFFcompare and was able to run Deseq2; however, most of my genes came up with Stringtie IDs and not ENMUSG ids (like below). Even with the ENMUSG IDs I didn’t get a gene name which is weird since after Deseq2 next to to the ENMUS ID the gene name is written…
I also tried annotating the genes it via Annotate Deseq2 but most of them came up as NA…
Hi @rjsoh
The merging step tries to combine known genes and transcripts (provided in the original GFF file) with predicted transcripts (by Stringtie). Anything that couldn’t be merged will still have the predictions from Stringtie.
So … maybe back up a bit more. How did that merging step go? These tools are very picky about format, but having data based on different versions of a genome assembly is another possible reason for annotation data that doesn’t merge well.
Thank you Jenna,
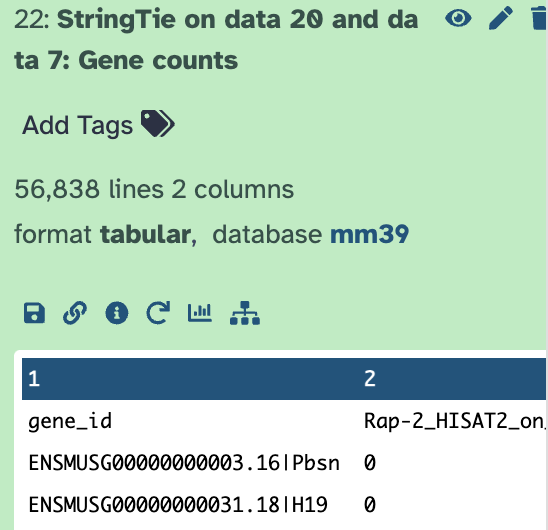
I will look into the earlier steps more. I’m also sharing my history showing that post stringtie using stringtie merge as my GTF I get different gene counts…
This dataset is comparing seq data from a previous experiment and a current one. I started with the fastgz files and went through the entire process rather than just using the HISAT bam files so I’m sure these were all mapped to the same genome etc…Sorry this is really confusing to me because I’ve analyzed this dataset before with no problem.
Thanks @rjsoh – the history helps.
Everything seems to be based on the mm39 assembly, and your merged annotation has the attributes I would expect, including the incorporated known attributes.
You could provide a two column mapping file to the counting tools instead of a GTF file to control which of those attributes are being used? Just watch out for any of the replaced values not being 1-many for gene-transcript.
This tutorial might be interesting for you too. Hands-on: Genome-wide alternative splicing analysis / Genome-wide alternative splicing analysis / Transcriptomics. Notice how the last tool in here consumes the predicted transcript (isoforms) from Stringtie merge, then maps all the differences back to the original set of genes in the reference annotation. The workflow in that tutorial isn’t perfect, but you can grab a copy of my suggested updates from this ticket and see the example output – including a Stringtie merge output that looks very similar to yours.
The point is that I think what you have is the expected output. Did you run through this before in Galaxy or somewhere else? How is the current behavior different? Maybe I am misunderstanding what you think is going on. Thanks!
Thank you this is so helpful.
I’m going to try the options you listed. I keep getting very few annotated genes by going through GFF compare and featurecounts…
I did run one of the samples through Galaxy before. I have 3 samples 5.5mM 25mM and Rapamycin. The 5.5mM and 25mM come from a previous sequencing experiment and I would like to compare my Rap sample back to the other two. I ran the rapamycin sample before in Galaxy with no issue and had no problem with Stringtie gene counts being the same number between all my samples and replicates. I think i’m just trying to avoid this error since my uneven number of gene counts post stringtie is maybe causing this error?
I’ve also just tried running stringtie with the Rapamycin triplicates since I’ve ran them on galaxy. I used the same encode, same genome, everything and it still gives me different number of gene counts, even when I exclude 5.5mM and 25mM.

I have had this exact same issue with different samples and it has yet to be resolved so please let me know if there are possible reasons for this.
The Stringtie tool is known to not count up the same features, and that is what this ticket is about addressing: Suggested update to workflow for differential-isoform-expression · Issue #5208 · galaxyproject/training-material · GitHub
That change is exploratory. If the underlying tool cannot be changed, then Galaxy cannot be changed. Why? It is the same tool everywhere: on the command line with R, or in Galaxy through the web application access.
Counting with Featurecounts instead is strongly recommended.
If you are not getting counts with Featurecounts, then that can point to an annotation GTF content issue of some sort. Or, a data issue (too many overlapping features, not grouped into the same gene?). Review the statistics that describe why matches where not counted up. Meaning, why are the reads not matching up to the features? Can something be changed? You can try to standardize the reference annotation data with gffread as one idea – see the options on the form for ways to re-cluster the features and maybe try those out?).
At the end of this, all of the count files must be based on the same set of gene IDs (or transcript IDs + gene IDs for the TMP usage) for DESeq2 to be able to process the DE analysis. The DESeq2 tool runs a check at the start – and if a different set of genes is detected (“different number of count file lines”), it will fail. That happens in Galaxy or anywhere else, so you don’t need to run the tool to discover this, you can just inspect the count files.
If something ran before, and is not working now, I would be interested in reviewing. I will need a history with the full prior run of Stringtie, and the new run of Stringtie. This can be for just one sample: the counts are different than they were before with the same inputs, same settings. I would be reviewing the tool version choices, and other technical differences. The data (BAM files + reference annotation) should be identical for this to be a true test about isolating a technical issue. Whoever can post that back is welcome too. I’ll review then post that example over to the issue ticket to provide more context to the development team. I’ve tried to do this myself, and I don’t get a difference, but of course could have missed something, or don’t have the correct type of data that triggers the difference.
Thanks and we can certainly follow up more here. ![]()
Hello,
Yes I was able to make this work by switching to FeatureCounts. Then when I annotated with Annotate Deseq2, i was able to get the gene names as well as the IDs.